Our Family Tree: a History of Our Family
Largest human family tree ever created retraces the history of our species
A new, enormous family unit tree for all of humanity attempts to summarize how all humans alive today relate both to one another and to our ancient ancestors.
To build this family unit tree, or genealogy, researchers sifted through thousands of genome sequences collected from both modern and aboriginal humans, as well as ancient human relatives, according to a new report published Thursday (Feb. 24) in the journal Scientific discipline. These genomes came from 215 populations scattered beyond the world. Using a computer algorithm, the team revealed distinct patterns of genetic variation inside these sequences, highlighting where they matched and where they differed. Based on these patterns, the researchers drew theoretical lines of descent between the genomes and got an thought as to which gene variants, or alleles, the common ancestors of these people likely carried.
In addition to mapping out these genealogical relationships, the team approximated where in the earth the common ancestors of the sequenced individuals lived. They estimated these locations based on the ages of the sampled genomes and the location where each genome was sampled.
Related: In photos: A nearly complete human being antecedent skull
"The way that we've estimated where ancestors live is, in particular, very preliminary," said first writer Anthony Wilder Wohns, who was a doctoral student at the Academy of Oxford'south Big Information Found at the fourth dimension of the study. Despite its limitations, the information still captured major events in human evolutionary history. For case, "nosotros definitely see overwhelming prove of the out-of-Africa event," meaning the initial dispersal of Man sapiens from East Africa into Eurasia and beyond, said Wohns, who is now a postdoctoral researcher at the Broad Institute of MIT and Harvard.
The method the researchers used "works well to refine known ancestral locations and, every bit sampling improves, information technology has the potential to identify currently unknown human being movements," Aida Andrés, an acquaintance professor in the Genetics, Evolution and Environment Department at the University College London (UCL) Genetics Establish, and Jasmin Rees, a doctoral candidate at the UCL Genetics Found, wrote in a commentary, likewise published in the journal Science on Thursday. So, in the futurity, when more data become available, such analyses could potentially reveal chapters of homo history that are currently unknown to us.
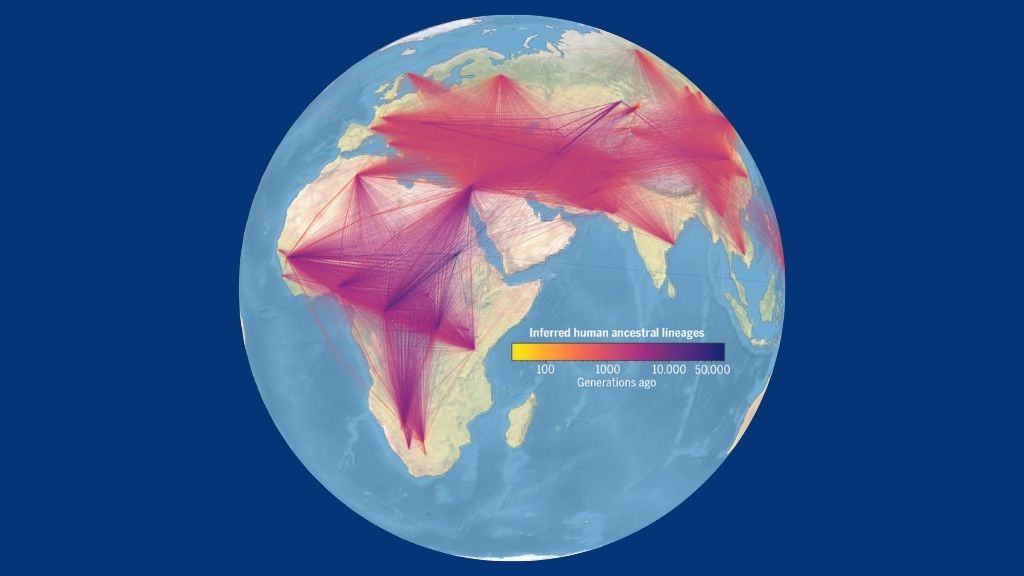
Edifice the human family tree
To build a unified genealogy of humanity, the researchers showtime pooled genomic data from several big, publicly available data sets, including the yard Genomes Project, the Human Genome Variety Project and the Simons Genome Diversity Project. From these information sets, they gathered about iii,600 high-quality genome sequences from modern-twenty-four hour period humans; "high-quality" genome sequences are those with very few gaps or errors, which accept been largely assembled in the right social club, according to a 2018 report in the journal Nature Biotechnology.
High-quality genomes from aboriginal humans were harder to come by, since Deoxyribonucleic acid from ancient specimens tends to be severely degraded, Wohns said. However, in earthworks through previously published inquiry, the team managed to find eight high-quality ancient hominin genomes to include in their tree. These included three Neanderthal genomes, one thought to be more than 100,000 years onetime; a Denisovan genome roughly 74,000 to 82,000 years old; and 4 genomes from a nuclear family unit that lived in the Altai Mountains of Russian federation about 4,600 years agone. (Neanderthals and Denisovans are extinct relatives of Human sapiens.)
In addition to these loftier-quality ancient genomes, the team identified more 3,500 additional, lower-quality genomes with meaning deposition, ranging from a few hundred to several thousand years old, Wohns said.
These degraded genomes did not factor into the main tree-building analysis, simply the team sifted through the fragments to see which isolated alleles could be identified in the samples. This piecemeal data helped the researchers ostend when different alleles first cropped up in the genealogical record, since the specimens that the genomes came from had been radiocarbon dated.
Ancient genomes provide a "unique snapshot of genetic diversity in the past," which can assistance reveal when and where a genetic variant first appeared, and how it spread thereafter, Andrés and Rees told Live Science in a joint statement. "Whilst this study does not integrate the low-quality ancient genomes into the building of the tree, using them to inform the historic period of variants within the tree is withal powerful for these means, and promises many exciting advances ahead."
Wohns and his colleagues used these information to double-check whether the lines of descent outlined in their family tree made sense, timing-wise — and, in almost cases, they did.
Related: Unraveling the human genome: 6 molecular milestones
"Information technology's very reassuring to see that … over 90% of the fourth dimension, we are being consistent with the samples that archaeologists can radiocarbon date," Wohns said. "But in that location are, yous know, v[%] or ten% of these genetic variants where we meet discordant estimates" every bit to when they first appeared, according to alien results from the archaeological record and the estimates made by their tree-building algorithm, he noted. In these cases, the squad adjusted their tree to reflect the timing that could be confirmed through radiocarbon dating, he said.
Although it'southward based on just a few thousand genome samples, the squad'south concluding family tree "actually captures quite a lot well-nigh the genealogy of all of humanity," Wohns said. Using the tree as a scaffold, the squad then conducted their geographical assay, to see when and where the theoretical ancestors of their sampled populations likely lived. From this, they not only found clear evidence of the out-of-Africa migration just as well uncovered potential evidence of interactions between Homo sapiens and now-extinct hominids, such as the Denisovans, he said.
For example, their results suggested that ancestors of modern humans could be found in Papua New Republic of guinea some 280,000 years ago, hundreds of thousands of years before the earliest known testify of modernistic human habitation in the region. That doesn't necessarily propose that H. sapiens actually occupied the expanse that long agone, "merely it does perhaps suggest that at that place's some genetic variation that is only constitute in that region, and indicates that there'south a really deep ancestry there that'south not plant elsewhere," he said.
Some of this unique ancestry may stem from modern humans breeding with Denisovans, as was besides suggested in a 2019 report in the journal Cell, which institute genomic prove of modern humans interbreeding with multiple Denisovan groups.
"The trees generated in this study will undoubtedly prove useful to those studying human evolution," only the methods and data used to construct said copse are "non without their limitations," Andrés and Rees wrote in their commentary. One limitation is that most genomic sequencing has been performed in Eurasian populations, so although the new report incorporated thousands of modern genomes, the data may not fully capture global genetic multifariousness, they told Live Science in an electronic mail. "Farther integration of under-represented populations would continue to tackle this limitation," they said.
"There'southward a lot of uncertainty in these estimates," Wohns said of the squad'south recent results. "Unless nosotros had the genome of everybody who e'er lived, and where and when they lived, that'southward the only style that we can get the truth." The team reconstructed human history as closely as they could given the data at mitt, simply with more genome samples and more than sophisticated software, the tree could definitely exist refined, he said.
Related: Photos: Looking for extinct humans in ancient cave mud
"The nice thing about the methods we've created is that they would piece of work with potentially millions of samples," Wohns said. "So, as we accept more data, we'll get ameliorate estimates."
Wohns said he'southward at present working to develop new car-learning algorithms to improve the team's estimates of where and when our ancestors lived. In a dissever project, he plans to employ the same tree-building method to improve understand the genetic footing of human disease. He aims to do this by pinpointing the origin point of disease-related alleles and then reconstructing how and when these gene variants spread through different populations.
The aforementioned tree-building method could also be used to trace the evolutionary history of other organisms, such equally honeybees or cattle, and even infectious agents, similar viruses, he added.
"The power and resolution of tree-recording methods promise to help clarify the evolutionary history of humans and other species," Andrés and Rees wrote in their commentary. "Information technology is likely that the almost powerful ways to infer evolutionary history going forrad will have their foundations firmly set in these methods."
Editor'southward note: This commodity was updated at 10 a.thou. on February. 25, 2022 with boosted comments from Aida Andrés and Jasmin Rees. The original article was posted at 7 a.one thousand. EST on the same twenty-four hours.
Originally published on Live Science.
Source: https://www.livescience.com/largest-human-family-tree-genealogy
0 Response to "Our Family Tree: a History of Our Family"
Post a Comment